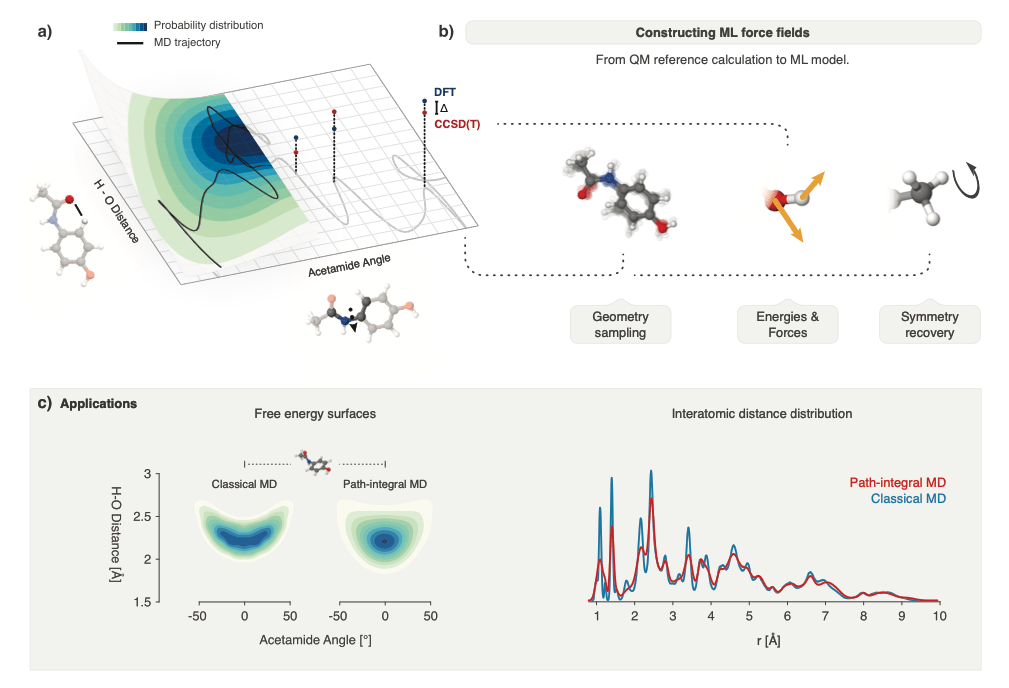
Researchers at Freie Universität Berlin and Rice University, Texas, reviewed recent ML (Machine Learning) methods for molecular simulation, with particular focus on (deep) neural networks for the prediction of quantum-mechanical energies and forces, coarse-grained molecular dynamics, the extraction of free energy surfaces and kinetics and generative network approaches to sample molecular equilibrium structures and compute thermodynamics.
To explain these methods and illustrate open methodological problems, they reviewed some important principles of molecular physics and described how they can be incorporated into machine learning structures. They identified and described a list of open challenges for the interface between ML and molecular simulation. (Rice University)
